Continuous Science: How Software Engineering Practices can solve Scientific Reproducibility
I published this article in Towards Data Science here
This article is about increasing reproducibility and productivity in computational biology by implementing modern software engineering practices.
I’m writing because it’s clear that the scientific community is becoming increasingly reliant on public software solutions — solutions that can be better supported by investing en-masse in evolving but exciting solutions in software engineering.
Introduction
Computational biologists spend more time benchmarking and assessing code than they do writing basic functionality.
Luckily for us software engineers face many similar problems and have developed a myriad of solutions for ensuring that software continues to perform well over its lifetime.
Continuous Integration and Continuous Delivery (CI/CD) is the culmination of these solutions, allowing software developers in the agile software development community to deliver solutions faster and more safely than ever before.
CI/CD is a culture and collection of practices which has an immense amount to offer the scientific community.
I’ve spent the past year working as a data scientist, building and benchmarking bioinformatic pipelines whilst working among experienced software developers for industry. During this time it has become clear to me that the introduction of CI/CD or an analogous system in the scientific computation world may have much to offer.
“Continuous Science” as I call it, offers a chance to improve reproducibility, reduce waste and accelerate our collective understanding throughout data driven science.
Current Challenges with Scientific Software
It is no secret that modern science is facing a reproducibility crisis and scientific software is no exception.
While reproducing a published paper is difficult in general, reproducing computation is especially difficult to achieve. Often, a scientist must retrieve all of the open source code, all of the data inputs and all of the data outputs of an existing solution before they can perform a comparison to their new proposal.
Not only is it a difficult and lengthy task to retrieve all of these resources, this is often not possible because of poor documentation of data, limited code sharing or version control and lack of expertise with prior solutions which may be implemented in different coding languages.
Challenges of reproducibility affect individuals acutely by slowing down their research and costing them valuable time that they must spend verifying software integrity.
The communal effects are more serious though, because this slows the progression of fields and hinders progress in important scientific domains like proteomics.
Unfortunately, all this means that people like myself who write scientific code can often spend more time trying to achieve valid comparisons between algorithms and code than building new and exciting solutions.
This is incredibly wasteful when we consider how much duplicated effort is occurring across labs globally.
Furthermore, if it’s hard for the tool-builders to compare, then it is likely almost impossible to believe that users such as biologists could be properly informed about the tools they are using.
Enter CI/CD and the development (pun intended) of Continuous Science.
What is Continuous Integration/Continuous Delivery and why do we need something like it?
CI/CD is a software-engineering culture built around automating the tasks of checking code automatically as new solutions are delivered. This means code can be updated all the time and engineers can be confident that it will perform well no matter how much it is changed.
I’ll explain this with an analogy — I love my Fitbit.
I use it to track metrics that are meaningful to my health, like my steps per day and my average heart rate. Sure, my Fitbit never forces me to exercise but it makes it really obvious when my good habits start dropping off and I need to get out more.
Automatically keeping track of the state of something like the health of your body or the health of your code as it changes makes it easier to maintain it and stay on track for your goals. Code health is real, as algorithms can lose compatibility with common input formats over time or become disorganized and buggy as they are edited. Like physical health, algorithms and tools can lose their edge over time if not cared for and monitored appropriately.
At Mass Dynamics, I’ve seen CI/CD applied by hosting servers with detailed instructions about how to run and test code every time we change it. By doing this we are able to automatically gain confidence that our code works at all times. If the checks weren’t automatic, we couldn’t possibly keep it as healthy.
By contrast, in the scientific community, we see a very different scenario.
While many scientific tools are awesome, the number of new algorithms/tools built each year creates not only work for the developer community through constant comparison and evaluation, but for users in distinguishing which tools are best for their needs.
One complication is that many tools can’t be run automatically and another is that they often take different inputs and produce slightly different outputs even for the same scientific problems, making comparisons difficult.
This means that confidence in new methods is slow and it is painstakingly gained by individual community members building benchmarking scripts (which are themselves published less frequently than the tool code itself). Often the results of these scripts are amazing papers which tell the story of an innovative solution but the efforts are duplicated (and often themselves hard to compare).
Going back to the health analogy, scientific software tools may be healthy, but we only know this because we pay for the occasional complex medical evaluation — or worse — surgery!
It shouldn’t be hard to know which scientific tools work. It shouldn’t be hard to know which scientific tools are more appropriate for a given user or problem.
So how do we solve this problem? How do we reduce the time and energy it takes for scientific developers to create and benchmark new solutions while at the same time making sure that consumers of that software can be confident about their use?
The Continuous Science Solution
In order to best articulate the solution I’m proposing, I’ve drawn a graphic that I hope will convey the modularity and elegance of the Continuous Science concept.
Figure 1 shows how datasets, algorithms and evaluation metrics can all be modularized and operate on a single server. This server could be a communal, transparent resource to show progress on a given computational challenge.
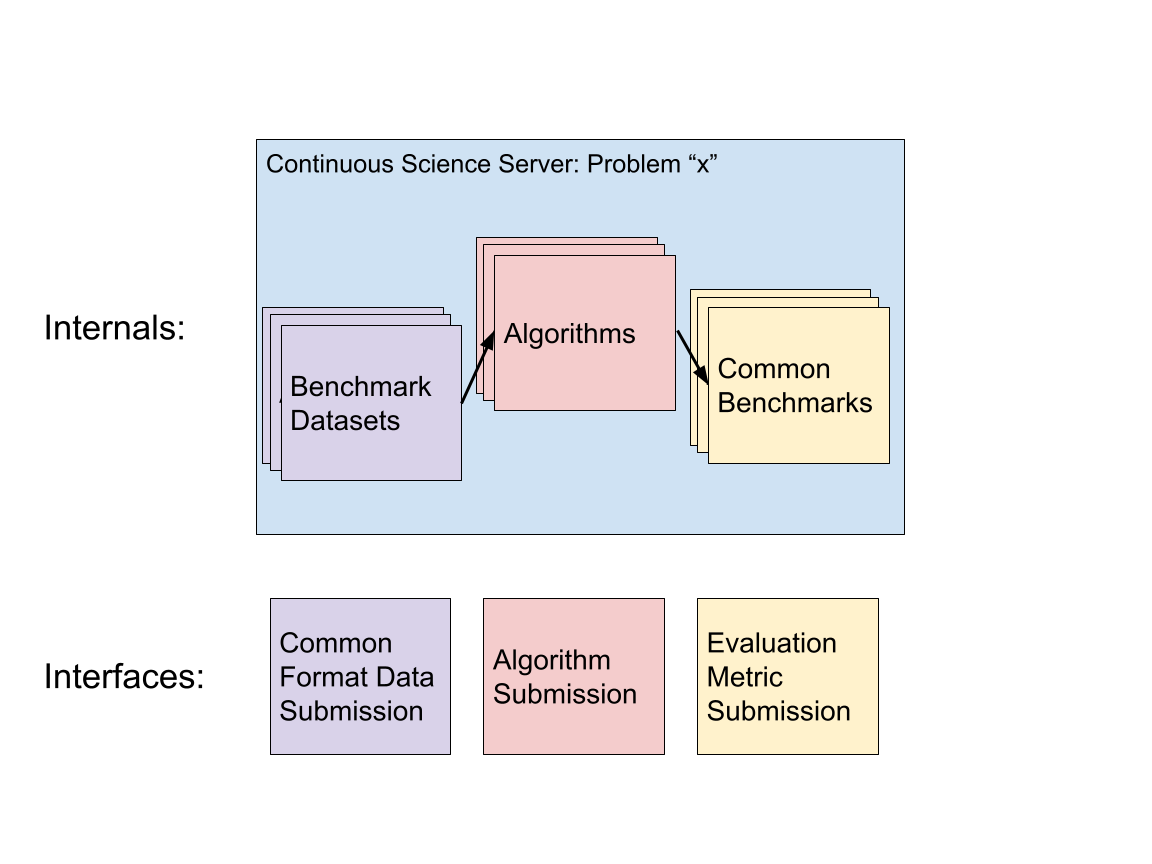 Figure 1: Original Figure by Author (Joseph Bloom)
Figure 1: Original Figure by Author (Joseph Bloom)
To elaborate, Continuous Science servers could work by sharing infrastructure and common data formats to allow algorithms to be compared to each other using common datasets and common benchmarks. Datasets, algorithms and benchmarks can all be submitted independently but are linked by a common problem. Performance of each algorithm across metrics and datasets should be publicly listed for each algorithm.
While CI/CD places emphasis on checking the health of the code, Continuous Science could assess algorithmic performance on common benchmarks and datasets that are transparent and accessible to the community.
If the community can come together and agree on common formats and benchmarks for algorithms that solve individual problems such as the protein inference problem, then a logical next step would be to set up communal servers that execute those benchmarks across all submitted algorithms and datasets.
The results could then be displayed in real time on a website that users of those solutions can browse, almost like a dashboard for the health of science.
Furthermore, one of the nice features of setting up a pipeline with modules for data, algorithms and evaluation metrics is that we can democratise the solutions. If a new solution performs well on a new, legitimate but previously un-anticipated metric or dataset type, these can be submitted to the tool and the results reviewed by all.
An example of a similar, awesome concept is Kaggle, a data science competition platform where people can submit problems with data and set benchmarks to compare to submitted answers. Data Scientists compete by submitting the results of different algorithms.
While “Kaggle” has a continuous flavour because each uploaded results set is automatically evaluated to check the quality of the solution, Continuous Science goes further in crowd sourcing test datasets and evaluation metrics.
Likely Challenges and Reason to Hope
Ideas are often far from practical at the outset. Considering some of the recent domains I’ve worked in, a number of immediate challenges jump to mind.
These are:
- The most popular solutions must be open source so that the community cares about the result of the comparison.
- Solutions must be automatable for the health check to be automatic.
- Solutions must share common inputs and outputs to enable fair comparisons.
Ultimately, to overcome these technological challenges we will need open and honest sharing and collaboration on the development of scalable solutions, contrary to the competitive culture which funding systems may foist upon us.
Despite how researchers are forced to compete for funding, there are many initiatives created by scientists working together to create more accessibility and transparency in science.
In particular, the drive towards “Open Science” as a solution to the reproducibility crisis is heartening. Efforts to create FAIR data (findable, accessible, interoperable and reusable) are making their way into highly technical domains such as Proteomics via papers such as this one and it makes it clear that there is global, communal goodwill and a shared desire towards making research less competitive and more collaborative.
A less competitive and more collaborative research community is exactly the kind that can share responsibility for maintaining the health of our algorithms via Continuous Science.
In Conclusion:
The reproducibility crisis is real, and we can take inspiration from awesome software development culture to help us solve it.
Reproducibility and accessibility of software in science can be addressed by a communal effort to track and compare the health of our algorithms via “Continuous science” servers, just like we track and maintain the health of our bodies with health devices and our code with CI/CD servers.
These common resources could then accelerate peer consensus, standardization in the same way that open source has affected the tech world more generally.
CI/CD culture brought into science is the way of the future. It will give scientific developers a communal resource to maintain and compare solutions which support non-computational scientists in making discoveries for the good of all.